Combined XN
Rietveld refinement with TOF data in GSAS-II
Introduction:
In these exercises you will use GSAS-II to refine the structure of Na2Ca3Al2F14 (NAC) from powder diffraction data collected at both APS and at the SNS both at room temperature. The data consists of one x-ray powder pattern collected on 11BM at λ=0.413909Å and two neutron TOF patterns from different wavelength bands (1.066Å and 2.066Å). The NAC sample has a minor (~10%) contamination from one of the starting materials (CaF2). NAC gives a very sharp diffraction pattern (sharper than SRM 660b) and thus can be used as a secondary standard (cubic I213, a~10.25Å). If you have not done so already, start GSAS-II.
Step 1. Import the phases
There are many ways to enter phase information into GSAS-II. Here we will simply extract it from an old GSAS .EXP file. To begin do Import/Phase/from GSAS .EXP file from the main GSAS-II data tree menu; a file dialog box will appear. Find the file TOF-CW Joint Refinement/data/NAC-2015A.EXP, select it and press Open. A popup window asking Is this the file you want?; it should obviously look like the right stuff so press Yes. Select NAC from the Read phase data popup and use the name NAC offered in the Edit phase name popup. After pressing OK, the General page will appear on the Phase Data for NAC window
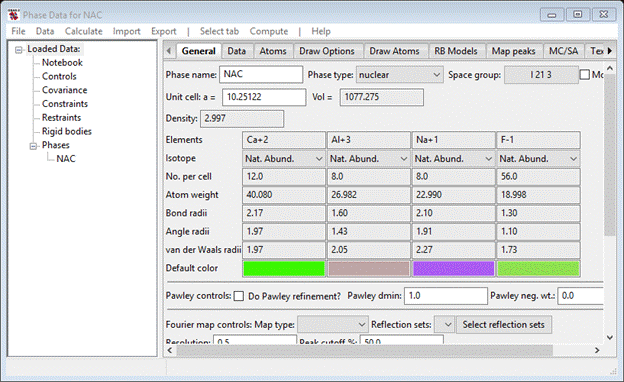
Now repeat the process (start with Import…) except select CaF2 from the Read phase data popup. Use the offered CaF2 for the name and press OK; the General page for Phase Data for CaF2 will be shown.
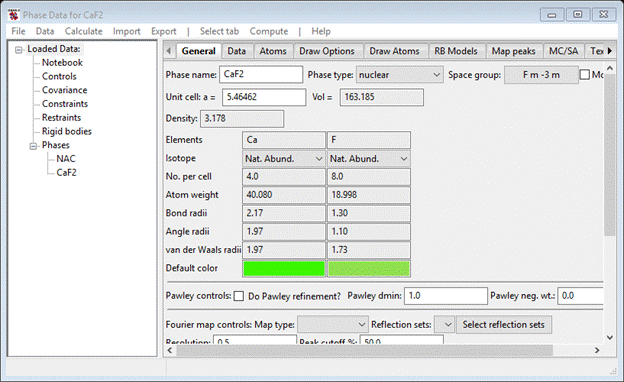
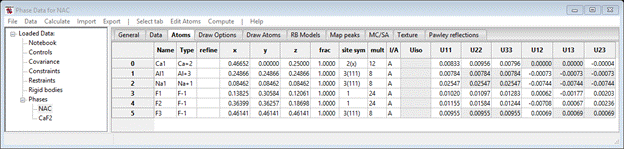
You can look at the various tabs to see what was extracted from the .EXP file. For example, NAC should have anisotropic thermal parameters.
Step 2. Import the data and set limits
There are three different data sets to be imported for this exercise. As they have different instrument parameter sets (more about this in a bit), they have to be imported one at a time. Multiple data files using the same instrument parameter file may be loaded together into GSAS-II. We will begin with the x-ray data; do Import/Powder Data/from GSAS powder data file from the main GSAS-II data tree menu. Again a file dialog box will appear; select TOF-CW Joint Refinement/data/11BM_NAC.fxye and press Open. Press Yes on the Is this the file you want? popup window. A new file dialog box will appear wanting the name of the corresponding instrument parameter file; select 11bm_gsas.prm and press Open. After a pause, the powder pattern will be shown on the plot and a popup window Add to phase(s) will appear. Select both NAC and CaF2 to connect them both to this data and press OK. Next select Limits from the GSAS-II data tree under the PWDR 11BM entry; a small data window will appear
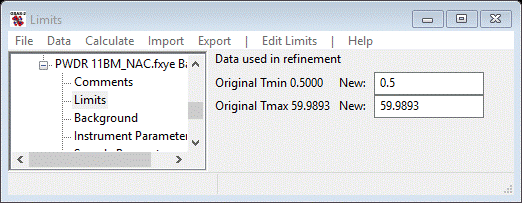
Change the Tmin changed to 2.5 and pick a clear spot in the pattern (zoom in beforehand) somewhere around 2Θ=32°. Use the right mouse button with the cursor on a data point to do this. A vertical red dashed line will appear where the upper limit is set. All zoom buttons must be off to do this pick.
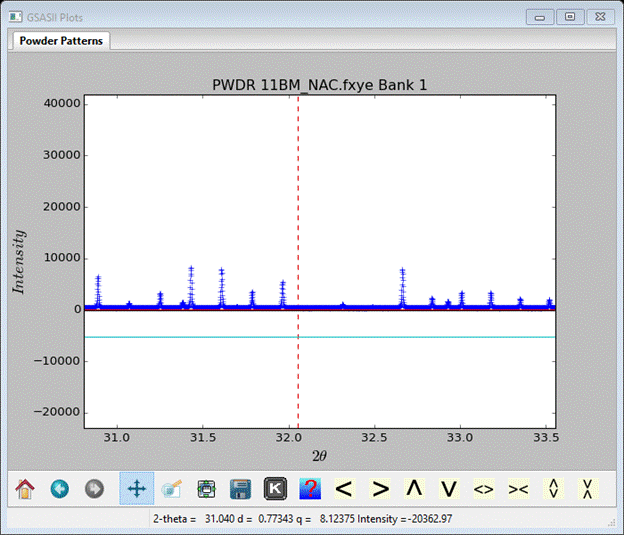
My pick is shown and the Limits window is something like

Now we’ll import the 1st TOF powder pattern; do the same Import command as above and pick PG_22048.gsa. The Is this.. window will show

Note that this is the 1.066A time frame. Press Yes; the instrument parameter file dialog box will appear but nothing seems suitable.
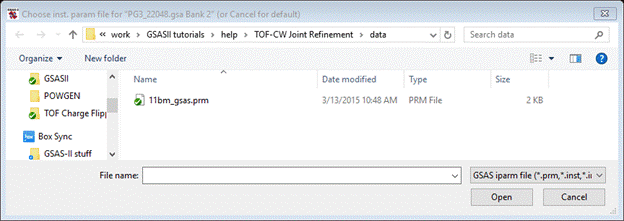
Select from the GSAS iparm file pull down the next entry, GSAS-II iparm file (*.instparm); the file dialog box will show two new & different instparm files.
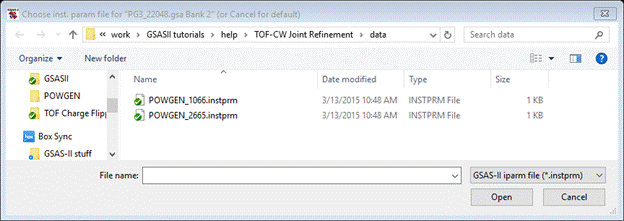
Select the 1066 one (for the 1.066A frame) and press Open. Again you will see the Add to phase(s) popup; select both phases and press OK. Next select Limits; a small Data window will appear
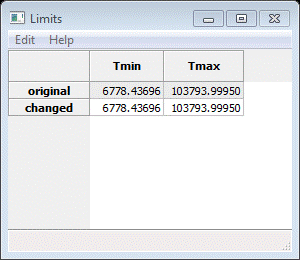
And the plot will show two vertical dashed lines (you may have to press the ‘Home’ button to make it show full scale). The upper one is ok where it is but the lower one must be moved to a more reasonable place. Zoom in on the low TOF end of the data. Find a place where there are still peaks and put the lower limit into a convenient gap. I chose a point where d~0.52A.
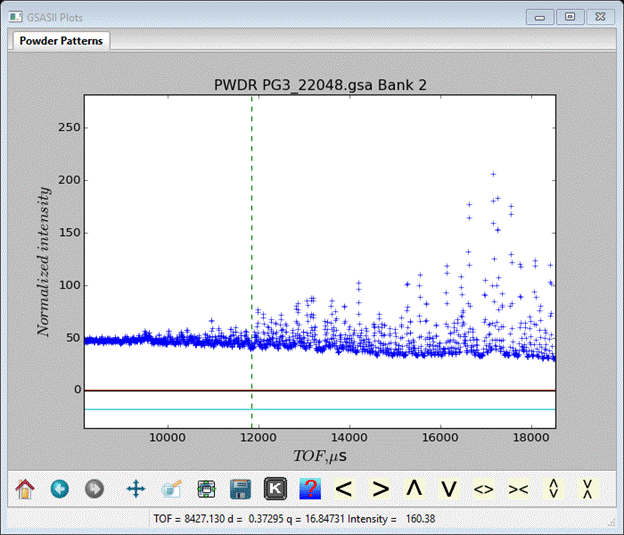
Finally, we’ll now load the third data set. Do Import/Powder data… as before and select PG3_22049.gsa. Press Open and note that this data is from the 2.665A frame. Again you’ll have to select GSAS-II iparm file (*.instparm) from the pulldown; pick the POWGEN_2665.instprm file and press Open. Again you will see the Add to phase(s) popup; select both phases and press OK. As this is the longer d-spacing portion of the pattern, peaks are clearly visible over the entire range of TOF so we need not change the limits. This finishes the import portion of the exercise and is a good time to save the project; do File/save as… from the main GSAS-II data tree menu. I named it NX on NAC.
Step 3. Initial Rietveld refinement
By default, a simple 3 term Chebyschev background and the histogram scale factors are automatically set for refinement. Select Controls from the GSAS-II data tree
These defaults are ok so do Calculate/Refine. A progress bar will appear that tracks the residuals for each data set; finally a result popup will show the overall Rw (I got 51%). Press OK to load the results. The fit to the x-ray data is quite poor; a zoom in that shows a NAC and a CaF2 peak shows the major problem namely the relative amounts of each are wrong. The NAC peaks also appear to be sharper than what is calculated.

The neutron patterns have a similar problem and the 1st one could use more background terms. Go to the Background item under PWDR PG3_22048.gsa Bank 1 and change the No. coeff. to 6. Do the same for the other neutron pattern. Next go to the Phases/NAC in the GSASII data tree and select the Data tab in the Phase Data window.

This contains a number of nonstructural parameters of interest for the NAC phase as seen in the 11BM powder pattern. These include Phase fraction, size and strain parameters as well as preferred orientation and macroscopic strain parameters perhaps associated with thermal expansion. There are similar sets of these parameters for NAC in the two neutron TOF patters and for CaF2 in all three patterns. The default values are normally reasonable starting points for a refinement; however the extreme sharpness of the 11BM NAC peaks suggest that the mustrain value is too high; change it to 100.0; we will probably not refine it. Then select the Phase fraction check box so it will be refined. We want these values and refinement flags (checkboxes used to determine the parameters that will be varied) to be used for the two neutron TOF patterns as well. Do Edit Phase/Copy Data; a popup box will allow you to select the data sets to be copied to. Set All of them and press OK; this will copy the values and refinement flags to the other two histograms.
Now select the CaF2 phase from the data tree; the Data page will be repainted. Select the PWDR 11BM… data set and only check the Phase fraction box. The default values are ok in this case. Then do Edit Phase/Copy Flags. This only copies the refinement flags and model choices, and is useful if you don’t want to copy the values.
Now the set of parameters include some background coefficients and 9 scale factors for two phases in three histograms (powder patterns) which is 3 too many for refinement (a singular matrix would result). We need to provide 3 constraints to reduce this. Select the Constraints item from the GSAS-II data tree; an empty tabbed window is displayed.

Select the Histogram/Phase constraint tab; the window is still empty. What we want to do is constrain the sum of the NAC and CaF2 phase fractions to be one for each histogram. GSAS-II will apply these constraints to the values during the refinement; unlike in older programs (GSAS, TOPAS & FULLPROF) we do not need to preset the values. To begin do Edit Constr./Add constraint equation from the Constraints menu; a popup window will appear
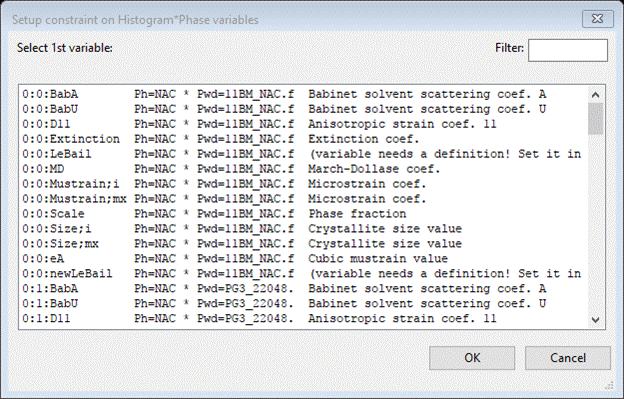
All the possible phase/histogram variables are listed. The prefix to each name indicates the phase (p) and histogram (h) for that parameter as p:h:name. The phase and histogram numbering begin with zero. The text following the name identifies the particular phase and histogram for each parameter along with a short description. Here we want the phase fraction for NAC in the 11BM data set (0:0:Scale); select it and press OK. A new popup window will appear with all the parameters that could be constrained to the 1st one chosen, here only Scale parameters
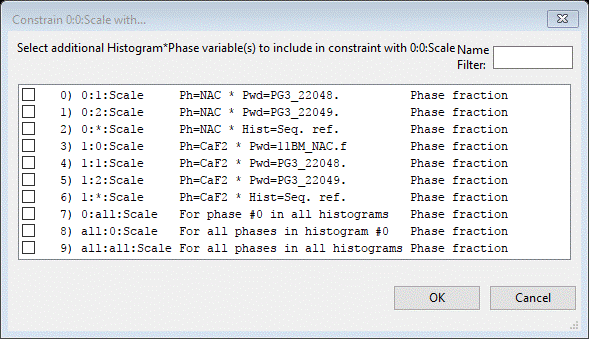
Here you can select one or more parameters to form the constraint. In this case we only want 0:0:Scale and 1:0:Scale so that the two phase fractions are constrained just for the 11BM data. Select (check box) for 1:0:Scale and press OK. The constraint will be shown on the Constraints window
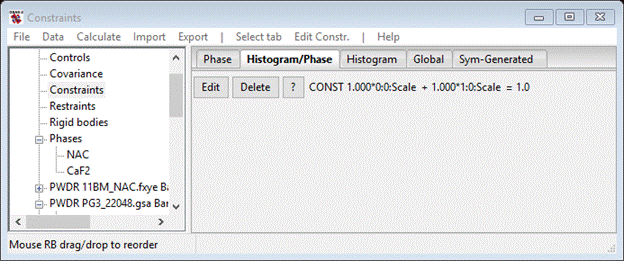
The constraint is that the sum of the phase fraction is one. You can edit the constraint changing multipliers and sum if need be, but that is not needed here. Now repeat the process for each of the neutron TOF histograms; constrain 0:1:Scale and 1:1:Scale for the PG3_22048 data and 0:2:Scale and 1:2:Scale for the PG3_22049 data. When done your Constraint page should look like.
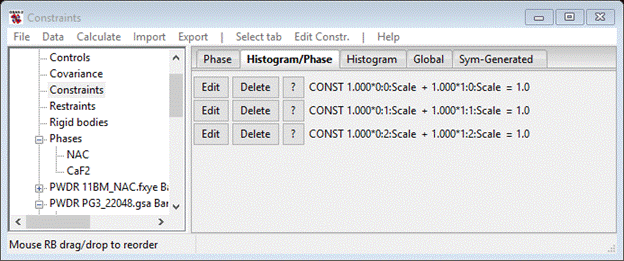
Then do Calculate/Refine; the result is much improved, I got Rw ~ 19%.
Step 4. Continued Rietveld refinement – nonstructural parameters
In looking over the patterns, the most apparent causes for the difference between the observed data and the calculated patterns depends on the type of data. For example, the 11BM result seems to suffer from broadening issues, namely the observed peak is narrower than the calculated one. That goes back to the fact that NAC is more crystalline than the materials used to calibrate that diffractometer (a mixture of SRM 640c and SRM 676a – Si & Al2O3, respectively). So it probably would be useful to refine U, V & W in the Instrument Parameters for PWDR 11BM_NAC.fxye; find this in the GSAS-II data tree and check the U, V & W Refine? boxes.

A common problem with Debye-Scherrer data collections is that the sample position is displaced slightly from the 2Θ axis. Select the Sample Parameters item for the 11BM data set.
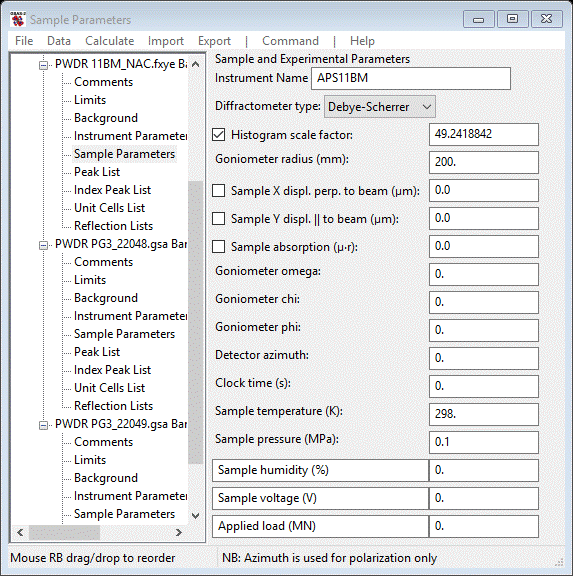
The Goniometer radius for 11BM is 1 meter; put 1000.0 for that value. The Sample X displ. parameter can be refined for a short angular scan (we only went to ~32° 2Θ. To do Sample Y displacement requires a scan covering >90° 2Θ. Check only the Sample X displ. box; the Histogram scale factor box was set by default.
We happened to start with a very good estimate for the NAC and CaF2 lattice parameters, but we should refine them both anyway. Select each phase in turn from the GSAS-II data tree and set the Refine unit cell box for each in the General page (shown by default).
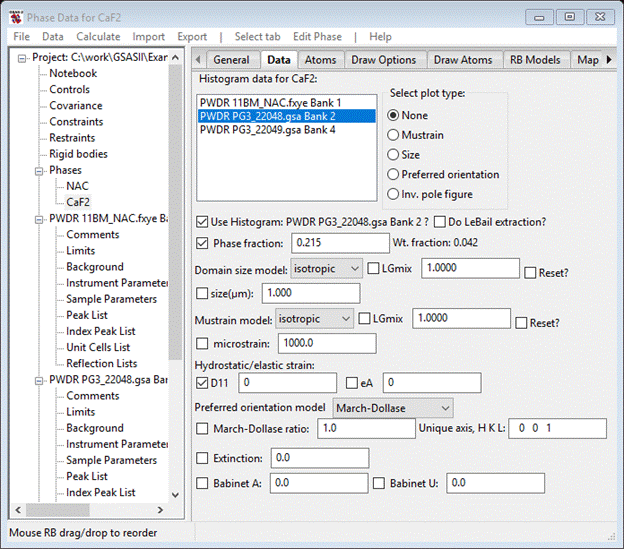
Looking at the two neutron TOF patterns seems to show mostly peak displacement problems in the λ=1.066Å one (higher resolution & sharper peaks). We have already selected to refine lattice parameters and the diffractometer constants (difC, etc.) were set from fitting standards (see the Calibration of a TOF powder diffractometer to see how this is done) so all that remains is a thermal expansion effect arising from small difference in data collection temperature between 11BM and POWGEN. The parameters for this are found on the Data tab for each phase. Select NAC from the GSAS-II data tree and select the Data tab. Select the PWDR PG3_22048.gsa Bank 1 data. Select the D11 box; your window should look like
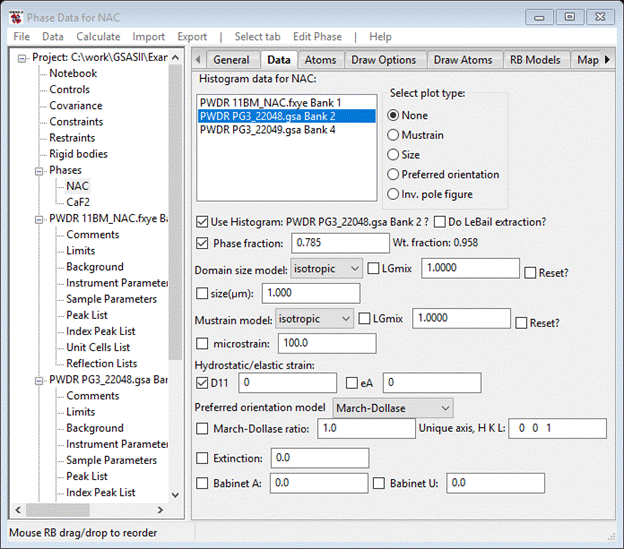
Do the same for CaF2; it will look like this when done.
Now do Calculate/Refine (perhaps twice) from the main GSAS-II data tree menu. The Rietveld refinement on 29 variables will proceed to give significant improvement. My Rw was 8.16% when done. The console shows some useful details on the progress of each refinement cycle.
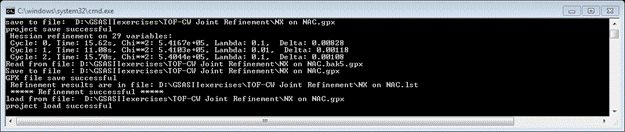
Notice that the Chi**2 (χ2) value has essentially leveled out and the Delta is close to zero for the last cycle. Delta is Δχ2/ χ2. Lambda is the Levenberg-Marquardt λ value; an indication of the damping used in the least squares. Here some damping was needed (minimum λ=10-5). Look at the Covariance matrix (item in tree).
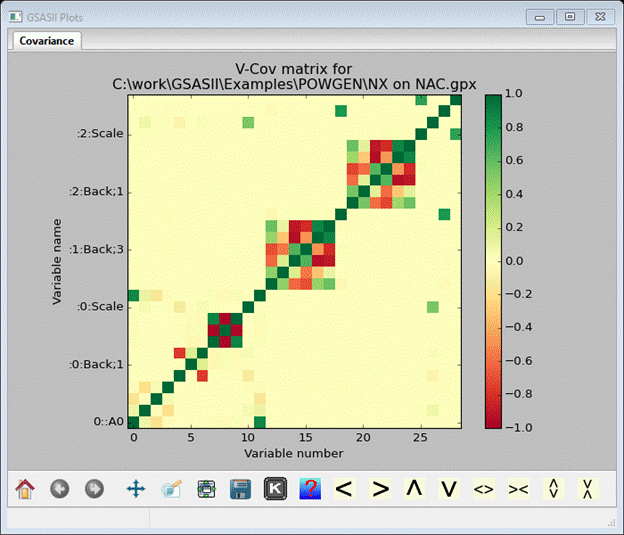
Move your cursor around the plot; it will show values and esds on the diagonal and covariances elsewhere. There are high covariance terms between the U, V & W profile coefficients for the 11BM data and some amongst the various background parameters; this probably necessitated the Levenberg-Marquardt damping.
Step 5. Structure refinement
Now that all the nonstructural parameters seem to be dealt with, we can include the crystal structure parameters in the refinement. Select the Phases/NAC entry from the GSAS-II data tree and choose the Atoms tab; the Atoms table will be shown.
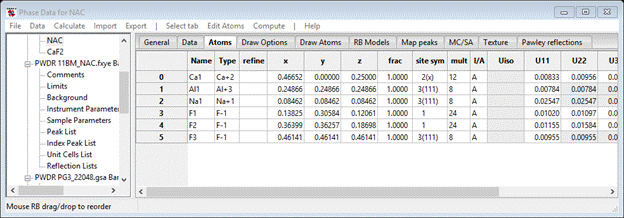
To refine parameters double click the refine column heading; a Refinement controls popup will show.
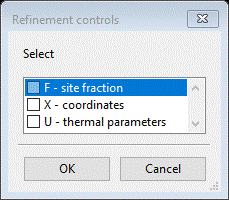
Select both X-coordinates and U-thermal parameters and press OK. Do not be concerned about atoms on special positions, GSAS-II will determine the refinement rules for these. The Atoms window will show the selected flags.
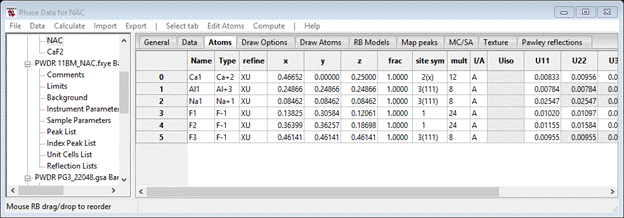
Do this for Phases/CaF2 although you need not bother setting X-coordinates as they are fixed in this structure.
Do Calculate/Refine twice to ensure convergence; my Rw=7.822%.
Step 6. Further work
You can now add parameters that haven’t been looked at earlier. These include size & mustrain for each phase in each data set. These should be made equivalent as appropriate; otherwise they may take on odd values in some cases. After including them, my residual was Rw=6.834% for 75 parameters and 40243 profile observations.